The discovery of antibodies can bypass processes such as immunization of animals and cell fusion to prepare monoclonal antibodies directly through phage display technology and by isolating human-derived antibodies to almost all antigens through a clonal selection of antibody fragments in a prokaryotic cell system, which has greatly progressed the field of antibody research.
The M13 phage is a group of filamentous phages collectively known as Ff phages. It can establish chronic infection in the host and continuously release new phages. Since Smith's successful integration of foreign DNA into the M13 phage chromosome in 1985, the role of M13 phages in antibody discovery has been gradually discovered. Since 1990, different antibody formats have been used to construct antibody display phage libraries. Generally, these antibody fragments are fused to the G3P of the M13 phage, and usually cloning a large number of genes encoding antibody fragments can generate large libraries of phage display antibodies from which many different antibodies can be selected.
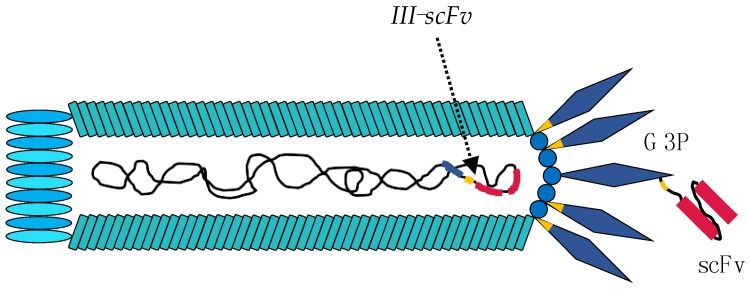 Fig 1. Engineering of the M13 phage for phage display experiments. (Ledsgaard, L., et al., 2018)
Fig 1. Engineering of the M13 phage for phage display experiments. (Ledsgaard, L., et al., 2018)
Since each phage particle typically contains three to five copies of G3P, the use of phage vectors has the potential to result in multivalent display of antibody–G3P fusion proteins (although proteolysis may result in the removal of a portion of the fused antibody). Therefore, phagemid vectors based on smaller "minimal plasmids" are a possible solution. Phagemid vectors are more typically used for library construction than phage vectors because higher transformation efficiency can be achieved to facilitate the construction of larger libraries.
The M13KO7 phage is a typical helper phage carrying a heterologous and low-copy origin of replication from the plasmid p15A inserted within the native M13 origin of replication. When the M13KO7 phage is present alone in the host bacterium (i.e., during the preparation of the helper phage), its replication is sufficient to produce high titers of the M13KO7 phage.
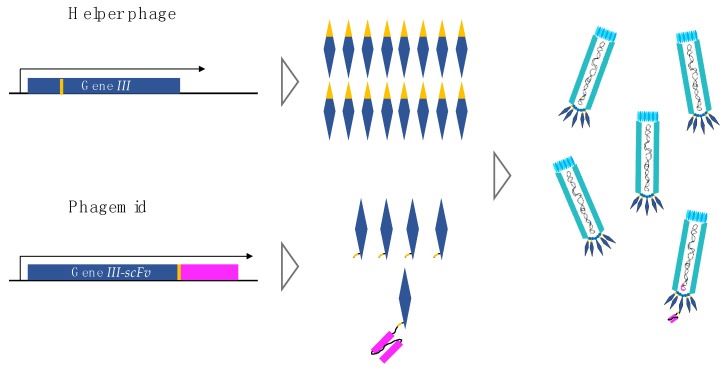 Fig 2. Gene III product from helper phage and gene III-scFv product from phagemid. (Ledsgaard, L., et al., 2018)
Fig 2. Gene III product from helper phage and gene III-scFv product from phagemid. (Ledsgaard, L., et al., 2018)
Phage display selection is a high-throughput method to discover specific antibodies against different antigens. The process consists of 5 main steps. First, the addition of the phage library. The library is added to the well or vial in which the antigen is presented. Antigen presentation can be achieved by direct coating (adsorption) or by a capture system (e.g., streptavidin-biotin). The second step involves binding, where the phage displaying the highest affinity antibody binds the epitope of the antigen, followed by washing the vial to remove the non-bound phages. After washing, the bound phages are eluted by enzymatic digestion using trypsin or other elution methods. When enzymatic digestion is employed, this step renders the bald phage non-infectious, thus improving the selection of still infectious antibodies displaying phage. In the fifth step, helper phages are added to amplify the eluted infectious phage in E. coli.
One of the most critical factors for the success of phage display selection experiments is also antigen presentation. Different antigen presentation methods have diverse pros and cons, and therefore the selection of the appropriate antigen presentation method needs to be considered in the context of the specific experimental situation.
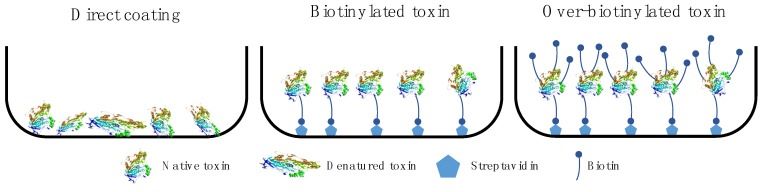 Fig 3. Antigen immobilization strategies. (Ledsgaard, L., et al., 2018)
Fig 3. Antigen immobilization strategies. (Ledsgaard, L., et al., 2018)
Phage display antibody libraries can be obtained from non-immunized (naïve) or immunized donors, depending on whether the donor from which the antibody gene was isolated and used to create the library. The benefit of the naïve libraries (usually IgM repertoire from non-immune donors) is that it can be used to discover a wide range of antigens, although the antibody fragments found may often have lower affinity than those found from immune sources. In phage-displayed antibody libraries from immune sources (from IgG repertoires), antibody fragments have greater affinity for the type of antigen used for immunization.
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.